US baby names dataset¶
This notebook gives an example of Active Anomaly Detection with coniferest and US baby names dataset.
Developers of conferest:
Konstantin Malanchev (LINCC Frameworks / CMU), notebook author
The tutorial is co-authored by Etienne Russeil (LPC)
Install and import modules¶
[1]:
%pip install 'coniferest[datasets]' pandas
Requirement already satisfied: pandas in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (2.3.0)
Requirement already satisfied: coniferest[datasets] in /Users/hombit/projects/supernovaAD/coniferest (0.0.11)
WARNING: coniferest 0.0.11 does not provide the extra 'datasets'
Requirement already satisfied: click in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (8.2.1)
Requirement already satisfied: joblib in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (1.5.1)
Requirement already satisfied: numpy in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (2.3.1)
Requirement already satisfied: scikit-learn<2,>=1 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (1.7.0)
Requirement already satisfied: matplotlib in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (3.10.3)
Requirement already satisfied: onnxconverter-common in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from coniferest[datasets]) (1.14.0)
Requirement already satisfied: scipy>=1.8.0 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from scikit-learn<2,>=1->coniferest[datasets]) (1.16.0)
Requirement already satisfied: threadpoolctl>=3.1.0 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from scikit-learn<2,>=1->coniferest[datasets]) (3.6.0)
Requirement already satisfied: python-dateutil>=2.8.2 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from pandas) (2.9.0.post0)
Requirement already satisfied: pytz>=2020.1 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from pandas) (2025.2)
Requirement already satisfied: tzdata>=2022.7 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from pandas) (2025.2)
Requirement already satisfied: six>=1.5 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from python-dateutil>=2.8.2->pandas) (1.17.0)
Requirement already satisfied: contourpy>=1.0.1 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (1.3.2)
Requirement already satisfied: cycler>=0.10 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (4.58.4)
Requirement already satisfied: kiwisolver>=1.3.1 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (1.4.8)
Requirement already satisfied: packaging>=20.0 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (25.0)
Requirement already satisfied: pillow>=8 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (11.2.1)
Requirement already satisfied: pyparsing>=2.3.1 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from matplotlib->coniferest[datasets]) (3.2.3)
Requirement already satisfied: onnx in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from onnxconverter-common->coniferest[datasets]) (1.17.0)
Requirement already satisfied: protobuf==3.20.2 in /Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages (from onnxconverter-common->coniferest[datasets]) (3.20.2)
Note: you may need to restart the kernel to use updated packages.
[2]:
import datasets
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import requests
from coniferest.pineforest import PineForest
from coniferest.isoforest import IsolationForest
from coniferest.pineforest import PineForest
from coniferest.session import Session
from coniferest.session.callback import TerminateAfter, prompt_decision_callback, Label
/Users/hombit/.virtualenvs/coniferest/lib/python3.12/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Data preparation¶
Download data and put into a single data frame
[3]:
%%time
# Hugging Face dataset constructed from
# https://www.ssa.gov/OACT/babynames/state/namesbystate.zip
dataset = datasets.load_dataset("snad-space/us-names-by-state")
raw = dataset['train'].to_pandas()
raw
Downloading data: 100%|██████████| 51/51 [00:01<00:00, 41.41files/s]
Generating train split: 100%|██████████| 6600640/6600640 [00:02<00:00, 2268886.44 examples/s]
CPU times: user 3.14 s, sys: 970 ms, total: 4.1 s
Wall time: 5.64 s
[3]:
| State | Gender | Year | Name | Count | |
|---|---|---|---|---|---|
| 0 | AK | F | 1910 | Mary | 14 |
| 1 | AK | F | 1910 | Annie | 12 |
| 2 | AK | F | 1910 | Anna | 10 |
| 3 | AK | F | 1910 | Margaret | 8 |
| 4 | AK | F | 1910 | Helen | 7 |
| ... | ... | ... | ... | ... | ... |
| 6600635 | WY | M | 2024 | Royce | 5 |
| 6600636 | WY | M | 2024 | Spencer | 5 |
| 6600637 | WY | M | 2024 | Truett | 5 |
| 6600638 | WY | M | 2024 | Wylder | 5 |
| 6600639 | WY | M | 2024 | Xander | 5 |
6600640 rows × 5 columns
Let’s load the data and transform it into a feature matrix where each column is the number (normalized by peak value) of US citizens that got this name in a given year. We apply few quality filter. We require the name to appear at least 10000 times over the full time range, this will prevent noisy data from names that are barely used.
Optionally, we use first few Fourier terms to better detect “bumps” and “waves” in the time-series.
[4]:
%%time
WITH_FFT = True
all_years = np.unique(raw['Year'])
all_names = np.unique(raw['Name'])
# Accumulate names over states and genders
counts = raw.groupby(
['Name', 'Year'],
).apply(
lambda df: df['Count'].sum(),
include_groups=False,
)
# Transform to a data frame where names are labels, years are columns and counts are values (features)
years = [f'year_{i}' for i in all_years]
year_columns = pd.DataFrame(data=0.0, index=all_names, columns=years)
for name, year in counts.index:
year_columns.loc[name, f'year_{year}'] = counts.loc[name, year]
# Account for total population changes
trend = year_columns.sum(axis=0)
detrended = year_columns / trend
# Normalise and filter
norm = detrended.apply(lambda column: column / detrended.max(axis=1))
filtered = norm[year_columns.sum(axis=1) >= 10_000]
if WITH_FFT:
# Fourier-transform, normalize by zero frequency and get power-spectrum for few lowest frequencies
power_spectrum = np.square(np.abs(np.fft.fft(filtered)))
power_spectrum_norm = power_spectrum / power_spectrum[:, 0, None]
power_spectrum_low_freq = power_spectrum_norm[:, 1:21]
frequencies = [f'freq_{i}' for i in range(power_spectrum_low_freq.shape[1])]
power = pd.DataFrame(data=power_spectrum_low_freq, index=filtered.index, columns=frequencies)
# Concatenate time-series data and power spectrum
final = pd.merge(filtered, power, left_index=True, right_index=True)
else:
# Use time-series data
final = filtered
final
CPU times: user 32 s, sys: 492 ms, total: 32.5 s
Wall time: 32.9 s
[4]:
| year_1910 | year_1911 | year_1912 | year_1913 | year_1914 | year_1915 | year_1916 | year_1917 | year_1918 | year_1919 | ... | freq_10 | freq_11 | freq_12 | freq_13 | freq_14 | freq_15 | freq_16 | freq_17 | freq_18 | freq_19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aaliyah | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.007815 | 0.011471 | 0.009205 | 0.002661 | 0.000384 | 0.003367 | 0.004686 | 0.002152 | 0.000062 | 0.000980 |
| Aaron | 0.045721 | 0.053748 | 0.062029 | 0.077329 | 0.074982 | 0.065564 | 0.066903 | 0.066013 | 0.065492 | 0.066256 | ... | 0.000203 | 0.000332 | 0.000003 | 0.000209 | 0.000003 | 0.000213 | 0.000325 | 0.000083 | 0.000084 | 0.000259 |
| Abbey | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.003283 | 0.002742 | 0.000650 | 0.000279 | 0.000762 | 0.000680 | 0.000231 | 0.000050 | 0.000016 | 0.000096 |
| Abbie | 0.359963 | 0.269810 | 0.306465 | 0.464630 | 0.215549 | 0.302426 | 0.349251 | 0.307615 | 0.197811 | 0.346344 | ... | 0.000792 | 0.000478 | 0.000180 | 0.000260 | 0.001956 | 0.000199 | 0.000055 | 0.000111 | 0.000089 | 0.000495 |
| Abbigail | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.002717 | 0.001827 | 0.001172 | 0.001199 | 0.001423 | 0.001123 | 0.000468 | 0.000081 | 0.000061 | 0.000128 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Zelma | 0.932474 | 1.000000 | 0.729411 | 0.723821 | 0.652394 | 0.669621 | 0.600106 | 0.576397 | 0.536196 | 0.576366 | ... | 0.012596 | 0.010955 | 0.009435 | 0.007556 | 0.005983 | 0.005520 | 0.005827 | 0.005979 | 0.005959 | 0.006366 |
| Zion | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.022560 | 0.009141 | 0.002182 | 0.006346 | 0.010988 | 0.008036 | 0.002212 | 0.001621 | 0.005530 | 0.006818 |
| Zoe | 0.009245 | 0.000000 | 0.003225 | 0.000000 | 0.001845 | 0.002542 | 0.004275 | 0.004373 | 0.002134 | 0.001221 | ... | 0.010147 | 0.009064 | 0.004659 | 0.001311 | 0.001038 | 0.002035 | 0.002939 | 0.003169 | 0.002656 | 0.002130 |
| Zoey | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.010202 | 0.010542 | 0.008716 | 0.005578 | 0.002431 | 0.000437 | 0.000107 | 0.000786 | 0.001459 | 0.001728 |
| Zuri | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.041216 | 0.032491 | 0.029391 | 0.026944 | 0.022914 | 0.018828 | 0.013810 | 0.008968 | 0.007011 | 0.007291 |
2337 rows × 135 columns
Plotting function
[5]:
def basic_plot(idx):
cols = [col.startswith('year') for col in final.columns]
all_years = [int(col.removeprefix('year_')) for col in final.columns[cols]]
if isinstance(idx, str):
counts = final.loc[idx][cols]
title = idx
else:
counts = final.iloc[idx][cols]
title = final.iloc[idx].name
plt.plot(all_years, counts.values)
# plt.ylim(-0.1, 1.1)
plt.title(title)
plt.xlabel('Year')
plt.ylabel('Count')
plt.show()
return
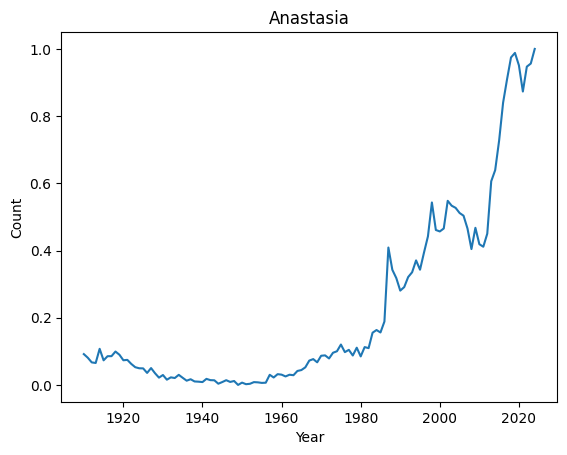
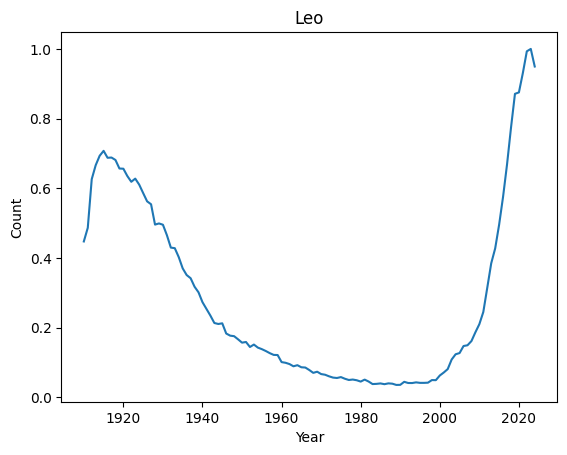
We can now easily look at the evolution of a given name over the years
[6]:
print(len(final))
basic_plot('Anastasia')
basic_plot('Leo')
2337
Classical anomaly detection¶
[7]:
model = IsolationForest(random_seed=1, n_trees=1000)
model.fit(np.array(final))
scores = model.score_samples(np.array(final))
ordered_scores, ordered_index = zip(*sorted(zip(scores, final.index)))
print(f"Top 10 weirdest names : {ordered_index[:10]}")
print(f"Top 10 most regular names : {ordered_index[-10:]}")
Top 10 weirdest names : ('Manuel', 'Marshall', 'Alfonso', 'Vincent', 'Margarita', 'Byron', 'Rudy', 'Benito', 'Peter', 'Ruben')
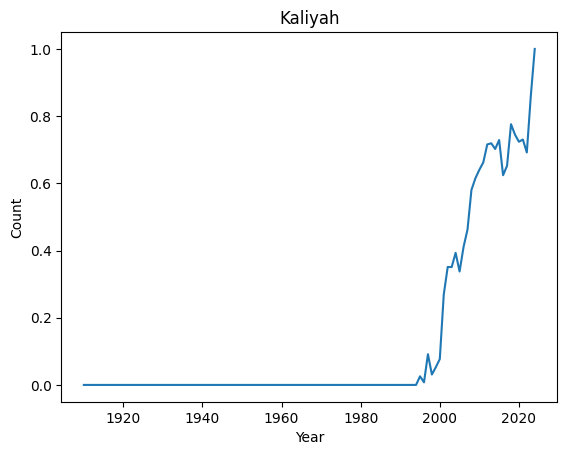
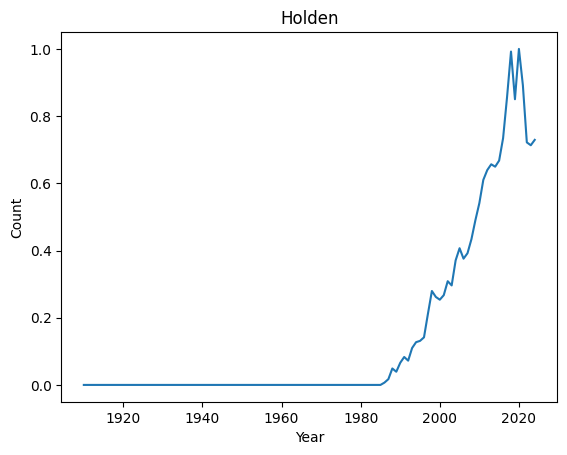
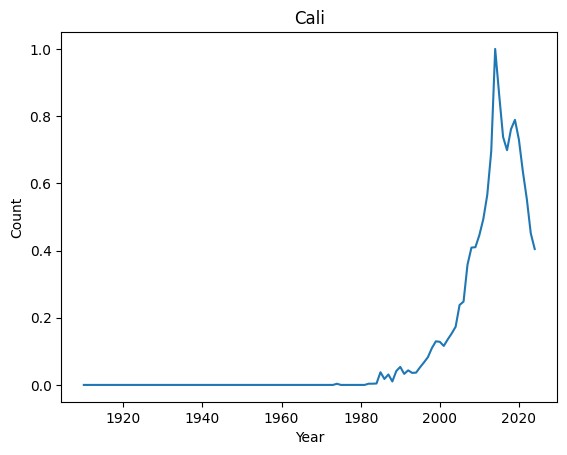
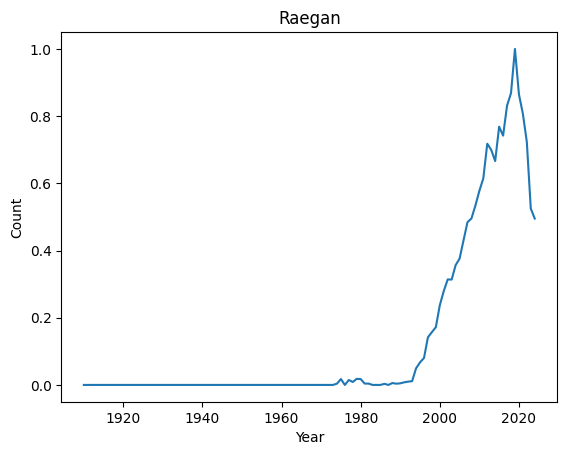
Top 10 most regular names : ('Kian', 'Jayla', 'Kenzie', 'Ibrahim', 'Teagan', 'Phoenix', 'Kaliyah', 'Holden', 'Cali', 'Raegan')
Let’s have a look at their distributions
[8]:
for normal in ordered_index[-4:]:
basic_plot(normal)
[9]:
for weird in ordered_index[:4]:
basic_plot(weird)
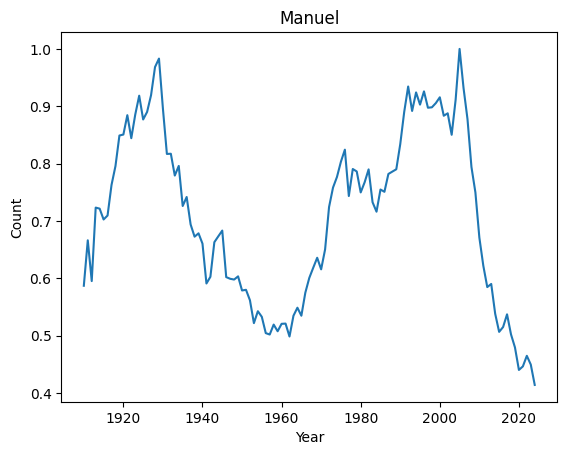
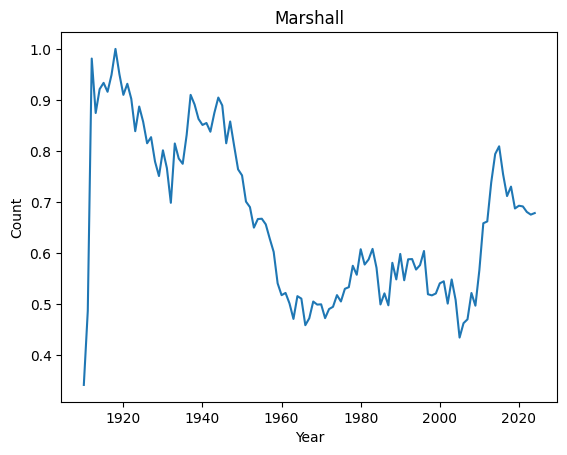
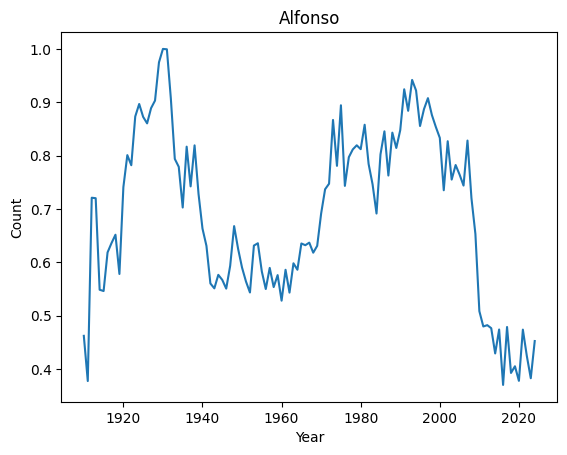
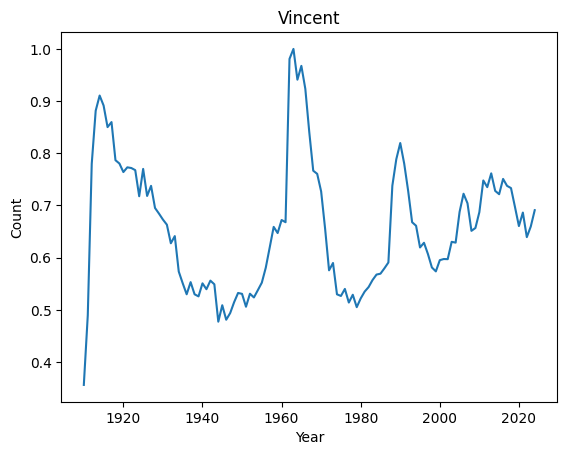
It seems that anomalies are either very localised peak or very recent trending names.
Active Anomaly Detection¶
First, we need a function helping us to make a decision.
Comment dummy decision function and uncomment interactive one¶
[10]:
# Comment
def help_decision(metadata, data, session):
"""Dummy, says YES to everything"""
return Label.ANOMALY
### UNCOMMENT
# def help_decision(metadata, data, session):
# """Plots data and asks expert interactively"""
# basic_plot(metadata)
# return prompt_decision_callback(metadata, data, session)
Let’s create a model and run a session
Let’s run PineForest and say YES every time we see a recent growth
[11]:
model = PineForest(
# Number of trees to use for predictions
n_trees=256,
# Number of new trees to grow for each decision
n_spare_trees=768,
# Fix random seed for reproducibility
random_seed=0,
)
session = Session(
data=final,
metadata=final.index,
model=model,
decision_callback=help_decision,
on_decision_callbacks=[
TerminateAfter(10),
],
)
session.run()
[11]:
<coniferest.session.Session at 0x12f081b20>
Wow! Good almost 100% of the behavior we were looking for
Now we can run it again and say YES every time we see a sharp peak
[12]:
model = PineForest(
# Number of trees to use for predictions
n_trees=256,
# Number of new trees to grow for each decision
n_spare_trees=768,
# Fix random seed for reproducibility
random_seed=0,
)
session = Session(
data=final,
metadata=final.index,
model=model,
decision_callback=help_decision,
on_decision_callbacks=[
TerminateAfter(20),
],
)
session.run()
[12]:
<coniferest.session.Session at 0x12f1930b0>
PineForest learns the profile interesting for the user and outputs it.
Try to change Pineforest hyperparameters and see how results change. Try different random seeds and number of trees.
[ ]: